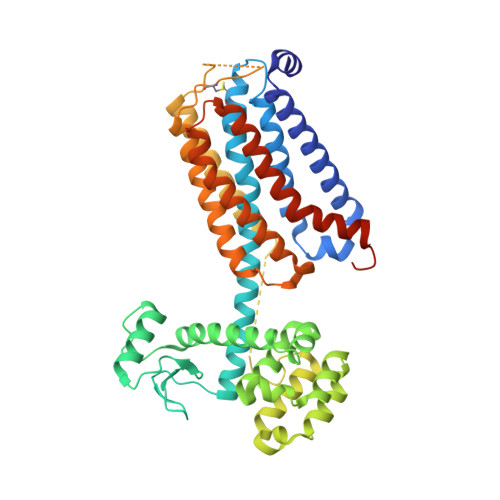
Fragment and Structure-Based Drug Discovery for a Class C GPCR: Discovery of the mGlu5 Negative Allosteric Modulator HTL14242 (3-Chloro-5-[6-(5-fluoropyridin-2-yl)pyrimidin-4-yl]benzonitrile).
Christopher, J.A., Aves, S.J., Bennett, K.A., Dore, A.S., Errey, J.C., Jazayeri, A., Marshall, F.H., Okrasa, K., Serrano-Vega, M.J., Tehan, B.G., Wiggin, G.R., Congreve, M.(2015) J Med Chem 58: 6653-6664
- PubMed: 26225459
- DOI: https://doi.org/10.1021/acs.jmedchem.5b00892
- Primary Citation of Related Structures:
5CGC, 5CGD - PubMed Abstract:
Fragment screening of a thermostabilized mGlu5 receptor using a high-concentration radioligand binding assay enabled the identification of moderate affinity, high ligand efficiency (LE) pyrimidine hit 5. Subsequent optimization using structure-based drug discovery methods led to the selection of 25, HTL14242, as an advanced lead compound for further development. Structures of the stabilized mGlu5 receptor complexed with 25 and another molecule in the series, 14, were determined at resolutions of 2.6 and 3.1 Å, respectively.
Organizational Affiliation:
Heptares Therapeutics Ltd. , BioPark, Welwyn Garden City, Hertfordshire AL7 3AX, U.K.